As before, this walk through picks up where the last left off. These scripts are meant to assist in separating combined fields, where unique data has been separated by 1 or more delimiters (separators). It also demonstrates pivoting data into a long format, so that it can be used in joining different data sets, based on like values, into one. I have also downloaded the .csv format of Trismegistos’ Geographical data dump to help locate names for the locations that did have matches in Pleiades. This is not a large download and requires no unzipping. They also offer the data in JSON format if you want to practice more with that.
I have the following objects loaded. These were generated in previous steps and saved as .rds files
pleiades_from_json_cleaned_tidied #Cleaned data from Pleiades JSON data
locations_5km #Locations within 5km of the original coordinates.
locations_master #My original locations list.
no_matches #Locations that did not find a match during the fuzzyjoin.What I wanted to do next was try to find locations that were not matched on the geo_inner_join(), and see if I could get attestations through Trismegistos (“TM” from now on) based on the name. I came to find out that was easier said than done.
The TM data was not tidy. There were multiple names, stored in many fields, across several columns. There was a composite variable (data created from combining 2 other fields): full_name that had additional info in it, so I could not ignore the column as redundant. The Latin names often had more than one known name, and were separated by a series of underscores, some extremely long. To illustrate this general untidiness, here is the TM record for one of my no_matches locations.

In the end, I wanted to have a long, tidy, list with a single row for each name instance, its Roman provincia, and country, all tied to its TM ID. That way I could perform a join on the city names and countries, and pull in more records, but also join the TM information to my final set.
Though these next sections are broken out, the script is meant to be one, long script, ran all at the same time. I did a series of separations using separate_wider_delim() to break out the data I needed, then a pivot_longer() to get everything in one long set. Finally, I did a little cleaning to get the names to a more standardized state. My location names would all be Romanized or in English, so I did not feel the need to include the Greek, Egyptian, or Coptic name translations. It is there if I ever want it, though.
# Start script
# Block 1
# Start big cleanup of tris data
tris_data_long_clean <- trismegistos_all_export_geo %>%
as_tibble() %>%
select(id,
provincia,
country,
name_latin,
name_standard,
full_name) %>%
separate_wider_delim(
full_name,
delim = " - ",
names = c("country_region_ext","city_ext"),
too_many = "merge",
too_few = "align_start") %>% Block 1
- Note:
trismegistos_all_export_geois the object loaded from the TM .csv file referenced above. From this, useselect()to pick only the necessary variables/columns. - Render the data set as a tibble for easier use/viewing.
- Start the first separation. It looks like there is only one initial break marked with space-underscore-space ( – ), which will be the delimiter in
delim =. The next steps are done inside the separation.- Specify the
full_namecolumn, and the delimiter. - Separate column into 2 columns:
"country_region_ext"and"city_ext". - In the event there is more or less data than can fit in 2 columns, what to do with those values . If there are more than 2 (
too_many), the remaining values are merged into the 2nd column with separators in tact. If there is 1 value (too_few), it will be placed incountry_ext, the start of the created columns.
- Specify the
# Block 2
separate_wider_delim(
country_region_ext,
delim = ",",
names = c("country_ext","region_ext"),
too_many = "merge",
too_few = "align_start") %>%Block 2
- Do another separation, just like above, this time breaking
country_region_extat the commas to get the country and regions alone.
#Block 3
mutate(city_ext = str_replace_all(city_ext, " \\[", "(")) %>% Block 3
- Mutate the
city_extcolumn usingstr_replace_all(). The inconsistent bracket vs. parenthesis make the next separation difficult. So, I want to replace left-brackets with left-parenthesis. The\\with the left-brackets are escape characters, and need to be used when working with certain characters inside R.* - * – stringr is a package inside the tidyverse. It’s a great set of functions that I use a lot for manipulating strings in R. I like to use this cheatsheet for help with understanding how different characters are handled inside package, and which need escaping.
# Block 4
separate_wider_delim(
city_ext,
delim = "(",
names = c("city_ext_1","city_ext_2"),
too_many = "merge",
too_few = "align_start") %>%Block 4
- Yet more separations. Just like 2 and 3 above, this time breaking
city_extat the parenthesis. This allows me to get at the appended information I discussed above.
# Block 5
separate_wider_delim(
name_latin,
delim = " - ",
names = paste0("tris_latin_", 1:25),
too_many = "merge",
too_few = "align_start") %>%
select(where(~ !all(is.na(.))),
-country_ext) %>% Block 5
- Some of these names have, like, 20+ delimiters (all consistent, thank God!), so I separate them into a large number of columns – more than I probably need. To do this, I use
paste0()to affix a “tris_” to the columns, then a sequence of 1-to-whatever I want. Here, 25 – seemed like plenty. This results in 25 columns namedtris_1throughtris_25,plenty of room for separation. - Because not every place will have 25 different names, those extra columns will be filled entirely with NA values. Here, the
select()function looks across all of my columns, and filters out those where all of the values in the column areNA. At the same time, I remove thecountry_ext, as it is truly redundant.
# Block 6
pivot_longer(!c(id, provincia, region_ext, country),
names_to = "source_seq",
values_to = "name") %>% Block 6
- Pivot this (now) very wide list into one long list using
pivot_longer(). Pivot everything exceptid, provincia, region_ext,andcountry. Save the column names into a new column called"source_seq"and the city names into"name".
# Block 7
mutate(name = str_trim(str_to_lower(name)),
region_ext = str_trim(region_ext),
name = if_else(name == "", NA, name),
country = str_to_lower(country),
name = str_replace_all(name, "\\)", "")) %>%
filter(!is.na(name)) %>%
select(-source_seq) %>%
distinct() %>%
relocate(name, .after = id)
#End scriptBlock 7
- Apply several mutations to clean up some of the columns:
- Trim the whitespace with
str_trim()innameandregion_ext(the separation process can generate a lot of it). I also converted the names to lowercase withstr_to_lower()for standardization. - Apply an
if_else()– if thenameis an empty field (represented by “”), then replace it with anNA, so it can easily be filtered out later. Else, leave whatever is in place. - Convert
countryto lower case. This should make the country names more standardized – important for joining later. - Replace the random right-parenthesis artifacts from the separation with blanks.
- Trim the whitespace with
- Filter out any
NAvalues in the name column. - Remove the
source_seqcolumn. It is not needed anymore. distinct()to condense all identical records into one, so each record is distinct.- Finally, move the
namecolumn so it is right after the id – easier for me to read.
Results
TM data before:
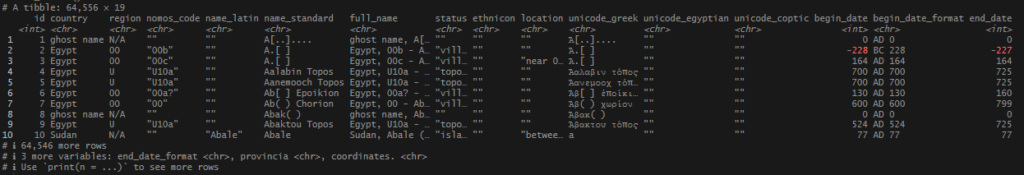
TM data after:

The final step to all of this was using this list to join the no_matches records to see if I could sweep in some of the TM sources where I did not have Pleiades. I also wanted to pull the TM ID’s from the Pleiades data and put them into my set. Since this script was so long, I stored the results in an object (tris_data_long_clean) and saved it as an .rds file for safe keeping.
# Save
saveRDS(tris_data_long_clean, paste0(objects_directory,"tris_data_long_clean.rds"))Let’s finally wrap this. Head over to part 2 to see how that was done.
Follow along script for parts 1 and 2 (in one file) are up on Github.
Leave a Reply