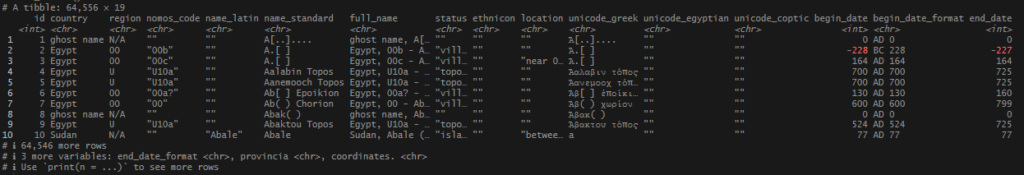
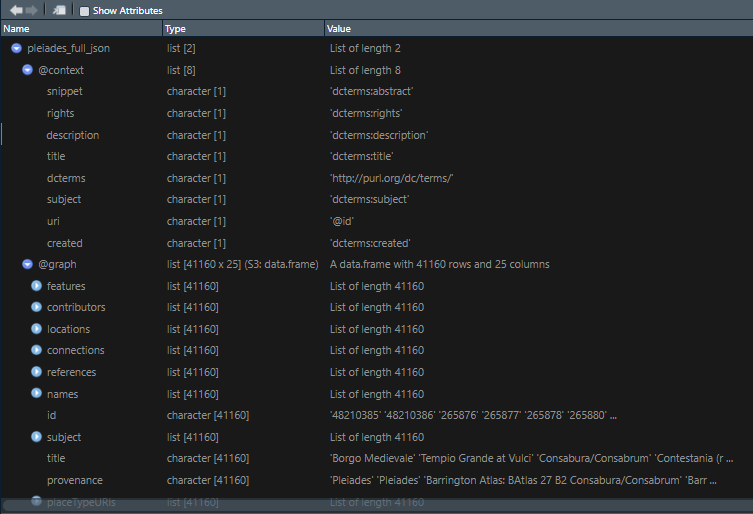
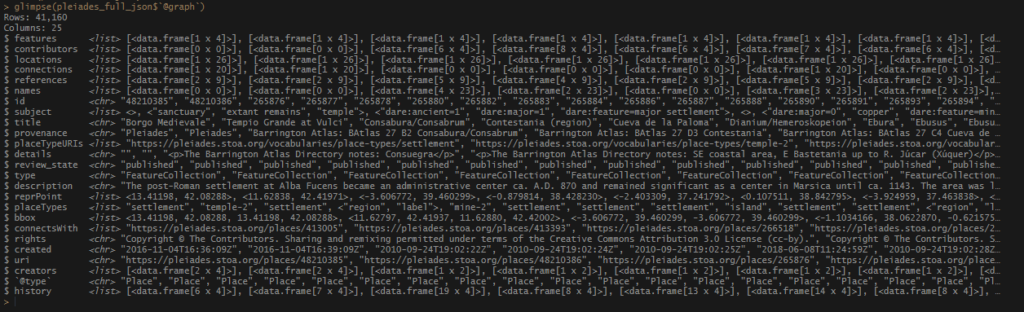
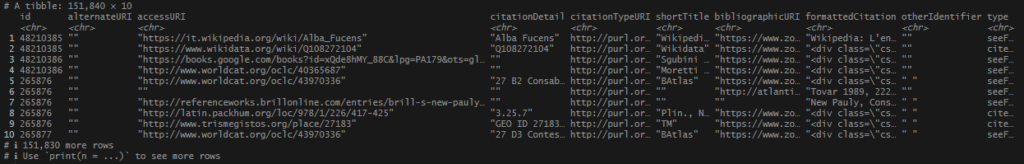
Once I had my long list of names, joining my no_matches set was very easy. I used an inner_join() and pulled in any record from tris_data_long_clean that matches my locations, both in name and in country. I was able to snag several more records.
tris_matches_2_no_matches <- no_matches %>%
mutate(ancientcity = str_to_lower(ancientcity),
country = str_to_lower(country)) %>%
inner_join(
tris_data_long_clean,
by = join_by(ancientcity == name,
country == country)) Steps
- Apply
str_to_lower()on bothancientcityandcountry. This helps standardize the name in case any random capitalizations exist, and assists in theinner_join(). inner_join()to match (exactly) on the name, and also the country, to ensure we have the right place. If there is no 1:1 match on name/country, then the record will be dropped. These locations will be re-joined later.
Results
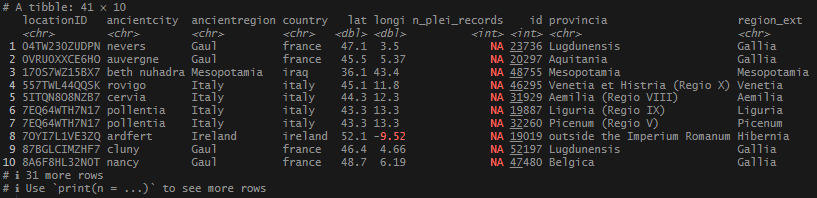
After this, I had 2 sets I wanted to bring together: locations_5km – the set joined by coordinates with Pleiades IDs; and then the new set generated from no_matches with TM IDs. I wanted to pull all of them together into one, tidy data frame. I remembered that Pleiades uses Trismegistos as one of their standard sources, and to keep things consistent, I decided to pull over the available TM ID’s from the Pleiades data. I also pulled over the Roman provincia, if it was available.
The following code, as in the post before, is broken up, but is actually one long script, meant to be run all at once.
# Start Script
# Block 1
# Bind the 2 data sets together by rows.
locations_plei_and_tris <- locations_5km %>%
rename("plei_id" = id) %>%
select(-long,
-lat) %>%
bind_rows(
tris_matches_2_no_matches %>%
rename("orig_lat" = lat,
"orig_long" = longi,
"tris_id" = id)) %>% Block 1
- With
locations_5km, I rename the ID column toplei_idto specify that it is the Pleiades ID. - Remove the long and lat column with
select()– I only want to use my original coordinate values. I will keep thedistance_from_originalvalue for reference. - Join the 2 sets:
locations_5kmandtris_matches_2_no_matches, by row withbind_rows(). This will “stack” the rows, combining like columns together, and pulling over whatever columns that are not in common between the 2 sets, and filling them with NA values. Within the bind, I clean up some column names withintris_matches_2_no_matches/
Next was a series of left_joins() to pull in the TM ID’s for the existing Pleiades ID’s I had and the provincia information for those ID’s.
# Block 2
left_join(
pleiades_from_json_cleaned_tidied$pleiades_references %>%
filter(grepl("trismegistos", accessURI, ignore.case = T)) %>%
mutate(tris_id_extract = str_extract(accessURI, "[0-9]+")) %>%
select(id, tris_id_extract),
by = join_by(plei_id == id)) %>% Block 2
- With a
left_join(), I access thepleiades_referencesinside the JSON list, a set I made previously. The next steps are done inside the join.filter()using agrepl()function. This will let you look at values in theaccessURIcolumn that contain the string “trismegistos”. I also applyignore.case =(just to be safe). This should pull all of the Trismegistos URLs.- Create a new column with
mutate()that will usestr_extract()to pull only the ID from the URL. The"[0-9]+"is a regex expression that tells the function to only extract any number 0-9, and any additional number after it. This gives me a nice neat column with only the TM ID intris_id_extract. - Select only the
tris_id_extractand the (Pleiades)id.
- Finally, join the 2 sets by the ID columns.
# Block 3
left_join(
tris_data_long_clean %>%
select(id,
provincia,
country) %>%
mutate(id = as.character(id)) %>%
distinct(),
by = join_by(tris_id_extract == id)
) %>% Block 3
- Apply another
left_join()withtris_data_long_clean(a set made in a previous step) to get the Roman provincia names. The next steps are also done inside the join.- Select only the
id,provincia, andcountrycolumns. - With
mutate(), convert theidto character format to facilitate the join. - Generate only distinct records.
- Select only the
- Finally, join the 2 sets by the ID columns.
# Block 4
mutate(trismegistos_id = coalesce(as.character(tris_id), tris_id_extract),
trismegistos_provincia = coalesce(provincia.x, provincia.y)) %>%
rename("country" = country.x) %>%
select(-provincia.x,
-region_ext,
-tris_id_extract,
-provincia.y,
-country.y,
-tris_id)
# End script.Block 4
- After the data was joined together, I have TM ID’s, in 2 columns, from the no_matches and from where I extracted the ID’s from the reference URL’s. I use
coalesce()to pull them together into one column:trismegistos_id. - Perform the same step on the provincia columns to create
trismegistos_provincia. rename() country.xtocountry.- Remove the redundant columns carried over.
Results
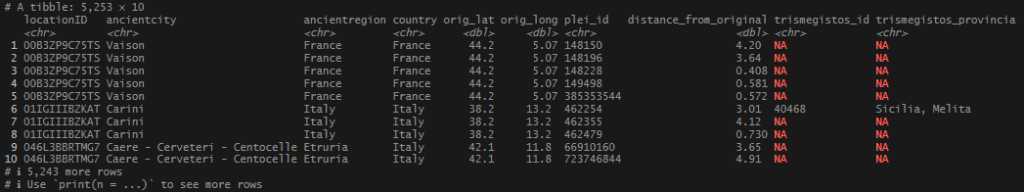
Getting close! I had all my distance-based location matches, and the extra name-based matches. I also had the TM ID’s for the Pleiades records that were pulled over on the geo_inner_join() from before. Just one more thing, pull in whatever of my original records where no location match was ever found.
locations_cited <- locations_master %>%
rename("orig_region" = ancientregion,
"orig_country" = country) %>%
left_join(
locations_plei_and_tris %>%
select(-ancientcity,
-ancientregion,
-country,
-orig_lat,
-orig_long),
by = join_by(locationID == locationID)) Steps
- In the original
locations_master, apply better names to reflect which source the region and country came from – assign withrename(). left_join()to thelocations_plei_and_trisobject that was just created. Inside the join, remove what would be redundant data. AnylocationIDsthat do not have a match will haveNAvalues in both theplei_idandtrismegistos_idfields.- Join the 2 sets by
locationID. Store in the objectlocations_cited.
Quick look at some of the records that didn’t come over. These will require some more research or fuzzy matching to sweep them up. Fortunately, there were only 109 orphaned records out of my original set of ~1,300.
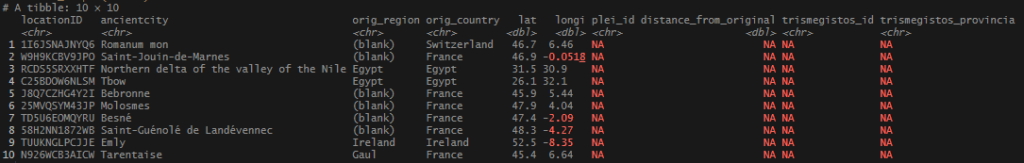
I also noticed that some of my locations independently pulled in a Pleiades ID and a TM ID, creating a duplicate row in some cases. I will have to figure out how suppress that in future iterations.
Despite that, I decided that this is really, really good locations data set, and that it would be OK to pause here and move onto something else. I was interested in taking one of my data sets, linking it to my locations data, and plotting it on a world map. Fortunately, I had a lot of other records tied to these locationID’s where I could do that.
Follow along script for parts 1 and 2 (in one file) are up on Github.